Generate single-cell images¶
Here, we are going to process the previously ingested microscopy images with the scPortrait pipeline to generate single-cell images that we can use to assess autophagosome formation at a single-cell level.
# Fix anndata>=0.12.0 forward slash restriction
import anndata._io.specs.registry as registry
original_write = registry.Writer.write_elem
registry.Writer.write_elem = (
lambda self, store, k, elem, dataset_kwargs=None: original_write(
self, store, str(k).replace("/", "|"), elem, dataset_kwargs=dataset_kwargs
)
)
import lamindb as ln
from collections.abc import Iterable
from pathlib import Path
from scportrait.pipeline.extraction import HDF5CellExtraction
from scportrait.pipeline.project import Project
from scportrait.pipeline.segmentation.workflows import CytosolSegmentationCellpose
ln.track()
Show code cell output
→ connected lamindb: testuser1/test-sc-imaging
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/xarray_schema/__init__.py:1: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
from pkg_resources import DistributionNotFound, get_distribution
→ created Transform('P35EpHHzarxP0000'), started new Run('MaUzrpv5...') at 2025-07-29 19:23:17 UTC
→ notebook imports: anndata==0.12.0 lamindb==1.10a1 scportrait==1.3.5
• recommendation: to identify the notebook across renames, pass the uid: ln.track("P35EpHHzarxP")
Query microscopy images¶
First, we query for the raw and annotated microscopy images.
input_images = ln.Artifact.filter(
ulabels__name="autophagy imaging", description__icontains="raw image", suffix=".tif"
)
The experiment includes two genotypes (WT and EI24KO) under two treatment conditions (unstimulated vs. 14h Torin-1).
Multiple clonal cell lines were imaged for each condition across several fields of view (FOVs) and imaging channels.
We’ll extract single-cell images from each FOV and annotate them with metadata including genotype, treatment condition, clonal cell line, and imaging experiment.
input_images_df = input_images.df(features=True)
display(input_images_df.head())
conditions = input_images_df["stimulation"].unique().tolist()
cell_line_clones = input_images_df["cell_line_clone"].unique().tolist()
FOVs = input_images_df["FOV"].unique().tolist()
Show code cell output
→ queried for all categorical features with dtype ULabel or Record and non-categorical features: (11) ['genotype', 'stimulation', 'cell_line_clone', 'channel', 'FOV', 'magnification', 'microscope', 'imaged structure', 'image_path', 'resolution', 'study']
| uid | key | genotype | stimulation | cell_line_clone | channel | FOV | magnification | microscope | imaged structure | resolution | study | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||
| 2 | Md4OouMExlWS2YfZ0000 | input_data_imaging_usecase/images/Timepoint001... | WT | 14h Torin-1 | U2OS lcklip-mNeon mCherryLC3B clone 1 | Alexa488 | FOV1 | 20X | Opera Phenix | LckLip-mNeon | 0.597976 | autophagy imaging |
| 3 | KKbVRkOjQ1jdA2fx0000 | input_data_imaging_usecase/images/Timepoint001... | WT | 14h Torin-1 | U2OS lcklip-mNeon mCherryLC3B clone 1 | Alexa488 | FOV2 | 20X | Opera Phenix | LckLip-mNeon | 0.597976 | autophagy imaging |
| 4 | CiQYTBNZrj0CPejK0000 | input_data_imaging_usecase/images/Timepoint001... | WT | 14h Torin-1 | U2OS lcklip-mNeon mCherryLC3B clone 1 | DAPI | FOV1 | 20X | Opera Phenix | DNA | 0.597976 | autophagy imaging |
| 5 | W6tzE7JNiM80Ruho0000 | input_data_imaging_usecase/images/Timepoint001... | WT | 14h Torin-1 | U2OS lcklip-mNeon mCherryLC3B clone 1 | DAPI | FOV2 | 20X | Opera Phenix | DNA | 0.597976 | autophagy imaging |
| 6 | YGiNq6DPfIEjtt9j0000 | input_data_imaging_usecase/images/Timepoint001... | WT | 14h Torin-1 | U2OS lcklip-mNeon mCherryLC3B clone 1 | mCherry | FOV1 | 20X | Opera Phenix | mCherry-LC3B | 0.597976 | autophagy imaging |
Alternatively, we can query for the ULabel directly:
conditions = ln.ULabel.filter(
links_artifact__feature__name="stimulation", artifacts__in=input_images
).distinct()
cell_line_clones = ln.ULabel.filter(
links_artifact__feature__name="cell_line_clone", artifacts__in=input_images
).distinct()
FOVs = ln.ULabel.filter(
links_artifact__feature__name="FOV", artifacts__in=input_images
).distinct()
By iterating through conditions, cell lines and FOVs, we should only have 3 images showing a single FOV to enable processing using scPortrait.
# Create artifact type feature and associated label
ln.Feature(name="artifact type", dtype=ln.ULabel).save()
ln.ULabel(name="scportrait config").save()
# Load config file for processing all datasets
config_file_af = ln.Artifact.using("scportrait/examples").get(
key="input_data_imaging_usecase/config.yml"
)
config_file_af.description = (
"config for scportrait for processing of cells stained for autophagy markers"
)
config_file_af.save()
# Annotate the config file with the metadata relevant to the study
config_file_af.features.add_values(
{"study": "autophagy imaging", "artifact type": "scportrait config"}
)
Show code cell output
→ transferred: Artifact(uid='voi8szTkmKPiahUA0000')
Process images with scPortrait¶
Let’s take a look at the processing of one example FOV.
# Get input images for one example FOV
condition, cellline, FOV = conditions[0], cell_line_clones[0], FOVs[0]
images = (
input_images.filter(ulabels=condition)
.filter(ulabels=cellline)
.filter(ulabels=FOV)
.distinct()
)
# Quick sanity check - all images should share metadata except channel/structure
values_to_ignore = ["channel", "imaged structure"]
features = images.first().features.get_values()
shared_features = {k: v for k, v in features.items() if k not in values_to_ignore}
for image in images:
image_features = image.features.get_values()
filtered_features = {
k: v for k, v in image_features.items() if k not in values_to_ignore
}
assert shared_features == filtered_features
# Get image paths in correct channel order
input_image_paths = [
images.filter(ulabels__name=channel).one().cache()
for channel in ["DAPI", "Alexa488", "mCherry"]
]
# Create output directory and unique project ID
output_directory = "processed_data"
unique_project_id = f"{shared_features['cell_line_clone']}/{shared_features['stimulation']}/{shared_features['FOV']}".replace(
" ", "_"
)
project_location = f"{output_directory}/{unique_project_id}/scportrait_project"
# Create directories
Path(project_location).mkdir(parents=True, exist_ok=True)
# Initialize the scPortrait project
project = Project(
project_location=project_location,
config_path=config_file_af.cache(),
segmentation_f=CytosolSegmentationCellpose,
extraction_f=HDF5CellExtraction,
overwrite=True,
)
# Load images and process
project.load_input_from_tif_files(
input_image_paths, overwrite=True, channel_names=["DAPI", "Alexa488", "mCherry"]
)
project.segment()
project.extract()
Show code cell output
Updating project config file.
INFO The Zarr backing store has been changed from None the new file path:
processed_data/U2OS_lcklip-mNeon_mCherryLC3B_clone_1/14h_Torin-1/FOV1/scportrait_project/scportrait.sdata
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
0%| | 0.00/25.3M [00:00<?, ?B/s]
4%|▎ | 944k/25.3M [00:00<00:02, 9.55MB/s]
39%|███▉ | 9.84M/25.3M [00:00<00:00, 55.8MB/s]
100%|██████████| 25.3M/25.3M [00:00<00:00, 85.6MB/s]
0%| | 0.00/3.54k [00:00<?, ?B/s]
100%|██████████| 3.54k/3.54k [00:00<00:00, 20.1MB/s]
0%| | 0.00/25.3M [00:00<?, ?B/s]
5%|▍ | 1.17M/25.3M [00:00<00:02, 12.3MB/s]
53%|█████▎ | 13.5M/25.3M [00:00<00:00, 81.4MB/s]
89%|████████▊ | 22.5M/25.3M [00:00<00:00, 86.7MB/s]
100%|██████████| 25.3M/25.3M [00:00<00:00, 80.5MB/s]
0%| | 0.00/3.54k [00:00<?, ?B/s]
100%|██████████| 3.54k/3.54k [00:00<00:00, 14.8MB/s]
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
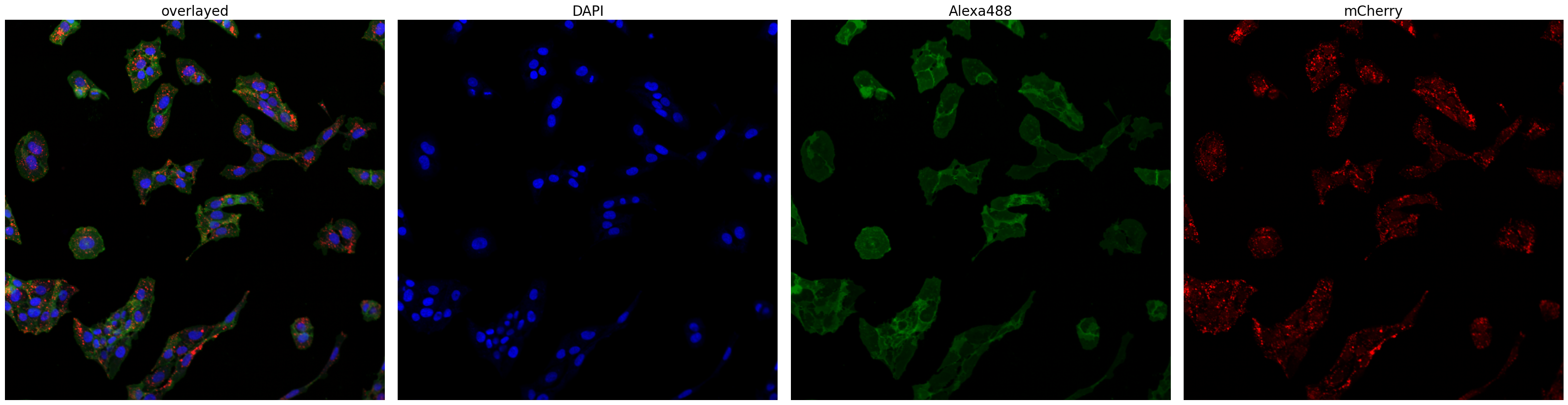
Let’s look at the input images we processed.
project.plot_input_image()
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
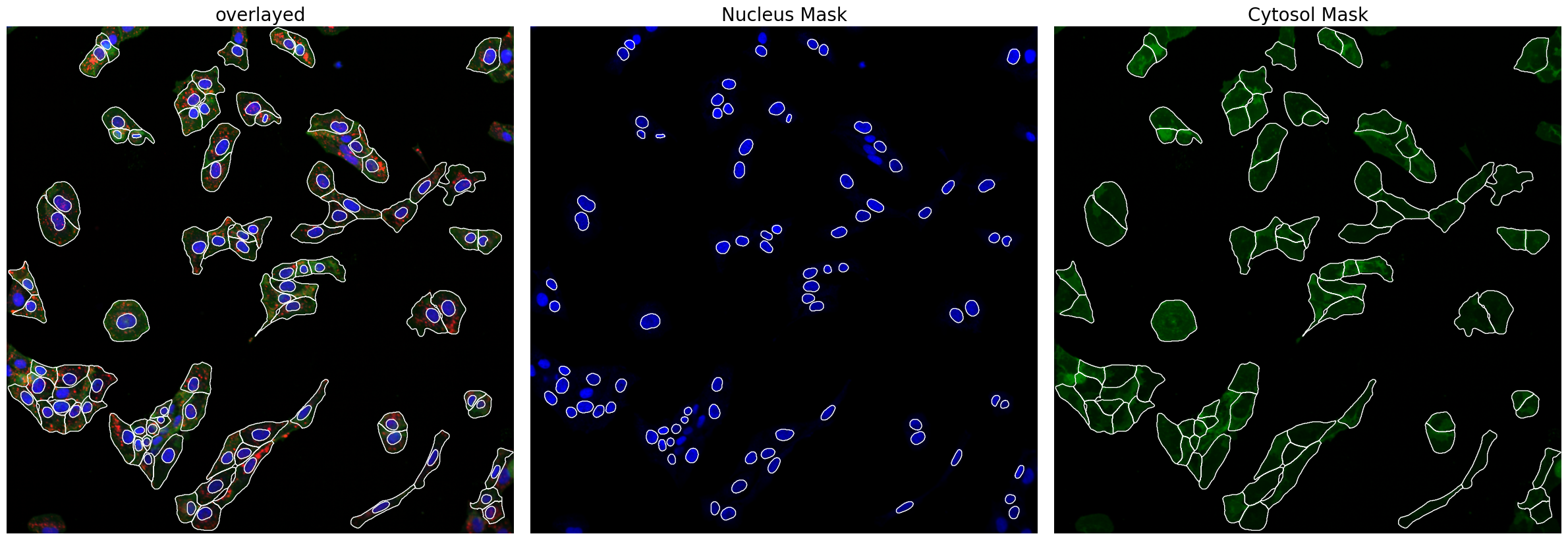
Now we can look at the results generated by scPortrait. First, the segmentation masks.
project.plot_segmentation_masks()
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
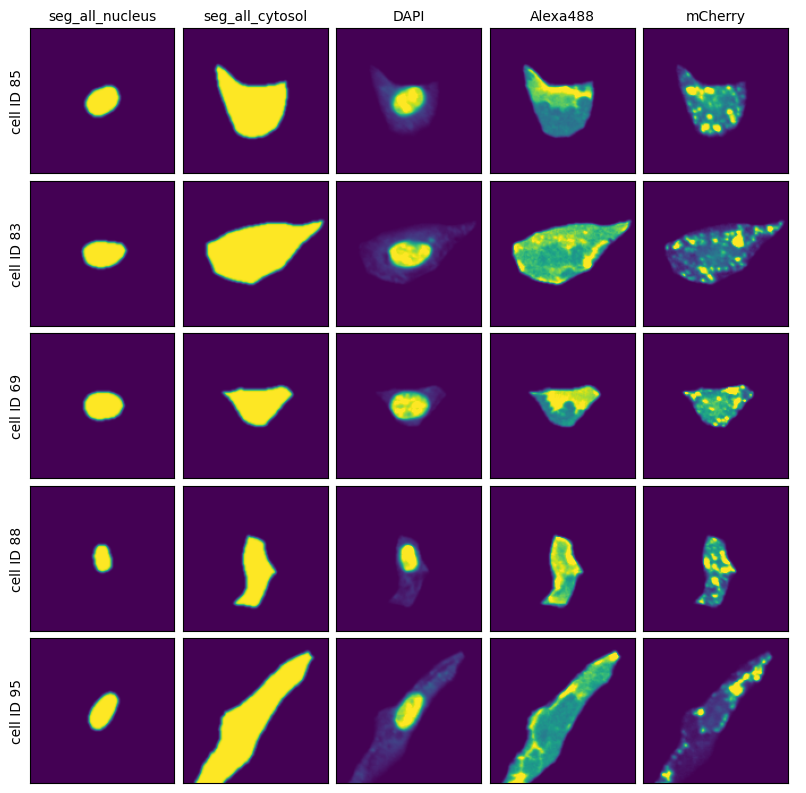
And then extraction results consisting of individual single-cell images over all of the channels.
# Fix anndata>=0.12.0 forward slash restriction
# The original code was: project.plot_single_cell_images()
from scportrait.plotting.h5sc import cell_grid
adata = project.h5sc
adata.uns["single_cell_images"] = {
k.split("|")[1]: v
for k, v in adata.uns.items()
if k.startswith("single_cell_images|")
}
cell_grid(adata, n_cells=5)
Save and annotate results¶
Now we also want to save these results to the instance.
ln.Artifact.from_spatialdata(
sdata=project.filehandler.get_sdata(),
description="scportrait spatialdata object containing results of cells stained for autophagy markers",
key=f"processed_data_imaging_use_case/{unique_project_id}/spatialdata.zarr",
).save()
Show code cell output
WARNING:ome_zarr.io:version mismatch: detected: RasterFormatV02, requested: FormatV04
INFO The SpatialData object is not self-contained (i.e. it contains some elements that are Dask-backed from
locations outside /home/runner/.cache/lamindb/Dl37ARo8eJlw7IfA0000.zarr). Please see the documentation of
`is_self_contained()` to understand the implications of working with SpatialData objects that are not
self-contained.
INFO The Zarr backing store has been changed from
processed_data/U2OS_lcklip-mNeon_mCherryLC3B_clone_1/14h_Torin-1/FOV1/scportrait_project/scportrait.sdata
the new file path: /home/runner/.cache/lamindb/Dl37ARo8eJlw7IfA0000.zarr
Artifact(uid='Dl37ARo8eJlw7IfA0000', is_latest=True, key='processed_data_imaging_use_case/U2OS_lcklip-mNeon_mCherryLC3B_clone_1/14h_Torin-1/FOV1/spatialdata.zarr', description='scportrait spatialdata object containing results of cells stained for autophagy markers', suffix='.zarr', kind='dataset', otype='SpatialData', size=6859499, hash='WGp8ykuoiN1VD_SRYwfgww', n_files=75, branch_id=1, space_id=1, storage_id=1, run_id=3, created_by_id=1, created_at=2025-07-29 19:24:37 UTC)
# Define schemas for single-cell image dataset
schemas = {
"var.T": ln.Schema(
name="single-cell image dataset schema var",
description="column schema for data measured in obsm[single_cell_images]",
itype=ln.Feature,
).save(),
"obs": ln.Schema(
name="single-cell image dataset schema obs",
features=[
ln.Feature(name="scportrait_cell_id", dtype="int", coerce_dtype=True).save()
],
).save(),
"uns": ln.Schema(
name="single-cell image dataset schema uns",
itype=ln.Feature,
dtype=dict,
).save(),
}
# Create composite schema
h5sc_schema = ln.Schema(
name="single-cell image dataset",
otype="AnnData",
slots=schemas,
).save()
Show code cell output
! you are trying to create a record with name='single-cell image dataset schema obs' but a record with similar name exists: 'single-cell image dataset schema var'. Did you mean to load it?
! you are trying to create a record with name='single-cell image dataset schema uns' but records with similar names exist: 'single-cell image dataset schema var', 'single-cell image dataset schema obs'. Did you mean to load one of them?
! you are trying to create a record with name='single-cell image dataset' but records with similar names exist: 'single-cell image dataset schema var', 'single-cell image dataset schema obs', 'single-cell image dataset schema uns'. Did you mean to load one of them?
# Curate the AnnData object
curator = ln.curators.AnnDataCurator(project.h5sc, h5sc_schema)
curator.validate()
# Save artifact with annotations
artifact = curator.save_artifact(
key=f"processed_data_imaging_use_case/{unique_project_id}/single_cell_data.h5ad"
)
# Add metadata and labels
annotation = shared_features.copy()
annotation["imaged structure"] = [
ln.ULabel.using("scportrait/examples").get(name=name)
for name in ["LckLip-mNeon", "DNA", "mCherry-LC3B"]
]
artifact.features.add_values(annotation)
artifact.labels.add(ln.ULabel(name="scportrait single-cell images").save())
! 5 terms not validated in feature 'columns' in slot 'var.T': '0', '1', '2', '3', '4'
→ fix typos, remove non-existent values, or save terms via: curator.slots['var.T'].cat.add_new_from('columns')
! no values were validated for columns!
! 11 terms not validated in feature 'keys' in slot 'uns': 'single_cell_images|channel_mapping', 'single_cell_images|channel_names', 'single_cell_images|compression', 'single_cell_images|image_size', 'single_cell_images|n_cells', 'single_cell_images|n_channels', 'single_cell_images|n_image_channels', 'single_cell_images|n_masks', 'single_cell_images|normalization', 'single_cell_images|normalization_range_lower', 'single_cell_images|normalization_range_upper'
→ fix typos, remove non-existent values, or save terms via: curator.slots['uns'].cat.add_new_from('keys')
! no values were validated for keys!
→ returning existing schema with same hash: Schema(uid='0000000000000000', name='single-cell image dataset schema var', description='column schema for data measured in obsm[single_cell_images]', is_type=False, itype='Feature', hash='kMi7B_N88uu-YnbTLDU-DA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, run_id=3, created_at=2025-07-29 19:24:37 UTC)
→ returning existing schema with same hash: Schema(uid='dt8Ycr2QAuoAq1IY', name='single-cell image dataset schema obs', n=1, is_type=False, itype='Feature', hash='Ordv-XQI-c0Bciy0UyH1_w', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, run_id=3, created_at=2025-07-29 19:24:37 UTC)
→ returning existing schema with same hash: Schema(uid='0000000000000000', name='single-cell image dataset schema var', description='column schema for data measured in obsm[single_cell_images]', is_type=False, itype='Feature', hash='kMi7B_N88uu-YnbTLDU-DA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, run_id=3, created_at=2025-07-29 19:24:37 UTC)
To process all files in our dataset efficiently, we’ll create a custom image processing function.
We decorate this function with :func:~lamindb.tracked to track data lineage of the input and output artifacts.
The function will skip files that have already been processed and uploaded, improving processing time by avoiding redundant computations.
@ln.tracked()
def process_images(
config_file_af: ln.Artifact,
input_artifacts: Iterable[ln.Artifact],
h5sc_schema: ln.Schema,
output_directory: str,
) -> None:
# Quick sanity check - all images should share metadata except channel/structure
values_to_ignore = ["channel", "imaged structure"]
first_features = input_artifacts.first().features.get_values()
shared_features = {
k: v for k, v in first_features.items() if k not in values_to_ignore
}
for artifact in input_artifacts:
artifact_features = artifact.features.get_values()
filtered_features = {
k: v for k, v in artifact_features.items() if k not in values_to_ignore
}
assert shared_features == filtered_features
# Create a unique project ID
unique_project_id = f"{shared_features['cell_line_clone']}/{shared_features['stimulation']}/{shared_features['FOV']}".replace(
" ", "_"
)
# Check if already processed
base_key = f"processed_data_imaging_use_case/{unique_project_id}"
try:
ln.Artifact.using("scportrait/examples").get(
key=f"{base_key}/single_cell_data.h5ad"
)
ln.Artifact.using("scportrait/examples").get(key=f"{base_key}/spatialdata.zarr")
print("Dataset already processed. Skipping.")
return
except ln.Artifact.DoesNotExist:
pass
# Get image paths in channel order
input_image_paths = [
input_artifacts.filter(ulabels__name=channel).one().cache()
for channel in ["DAPI", "Alexa488", "mCherry"]
]
# Setup and process project
project_location = f"{output_directory}/{unique_project_id}/scportrait_project"
Path(project_location).mkdir(parents=True, exist_ok=True)
project = Project(
project_location=project_location,
config_path=config_file_af.cache(),
segmentation_f=CytosolSegmentationCellpose,
extraction_f=HDF5CellExtraction,
overwrite=True,
)
project.load_input_from_tif_files(
input_image_paths, overwrite=True, channel_names=["DAPI", "Alexa488", "mCherry"]
)
project.segment()
project.extract()
# Save single-cell images
curator = ln.curators.AnnDataCurator(project.h5sc, h5sc_schema)
artifact = curator.save_artifact(key=f"{base_key}/single_cell_data.h5ad")
annotation = shared_features.copy()
annotation["imaged structure"] = [
ln.ULabel.using("scportrait/examples").get(name=name)
for name in ["LckLip-mNeon", "DNA", "mCherry-LC3B"]
]
artifact.features.add_values(annotation)
artifact.labels.add(ln.ULabel.get(name="scportrait single-cell images"))
# Save SpatialData object
ln.Artifact.from_spatialdata(
sdata=project.filehandler.get_sdata(),
description="scportrait spatialdata object containing results of cells stained for autophagy markers",
key=f"{base_key}/spatialdata.zarr",
).save()
ln.Param(name="output_directory", dtype="str").save()
Show code cell output
Feature(uid='YFnv2xG5veNe', name='output_directory', dtype='str', array_rank=0, array_size=0, branch_id=1, space_id=1, created_by_id=1, run_id=3, created_at=2025-07-29 19:24:38 UTC)
Now we are ready to process all of our input images and upload the generated single-cell image datasets back to our instance.
for condition in conditions:
for cellline in cell_line_clones:
for FOV in FOVs:
images = (
input_images.filter(ulabels=condition)
.filter(ulabels=cellline)
.filter(ulabels=FOV)
.distinct()
)
if images:
process_images(
config_file_af,
input_artifacts=images,
h5sc_schema=h5sc_schema,
output_directory=output_directory,
)
Show code cell output
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='Md4OouMExlWS2YfZ0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_Alexa488_zstack001_r003_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='0aoXxT857VvKAGo9UQo-8g', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='CiQYTBNZrj0CPejK0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_DAPI_zstack001_r003_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='YRjhTt2cBLq3BukWLUKC_w', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='YGiNq6DPfIEjtt9j0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_mCherry_zstack001_r003_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='zptjd_tR7P2Nw6mjUWbEjw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='KKbVRkOjQ1jdA2fx0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_Alexa488_zstack001_r004_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='Mm-DsaVdkMbTreUW_ipQBQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC), Artifact(uid='W6tzE7JNiM80Ruho0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_DAPI_zstack001_r004_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='9_LVWK08Z_D4c1mvIJlTOA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='uuh41FAHEz0ASL2N0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well01_mCherry_zstack001_r004_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='PSnZ2s1_uJa9G4_cc2HpNw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='yiIMSAddDWgLgki70000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_Alexa488_zstack001_r005_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='KUhrzT6sDoICDg9cGCUkmg', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='DEzw4QQAsjVZ010b0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_DAPI_zstack001_r005_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='N2xVJ8pQ9jWoywub8om_iQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='hbVyCGFARHU91Kax0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_mCherry_zstack001_r005_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='U2Hrdj8bUugok31k_uqEuw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='2ie2Kjzn1O7UYhuq0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_Alexa488_zstack001_r008_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='xkGVufN-7DDAZOeh4h7LpA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='fOQSb7JCK67aeN6a0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_DAPI_zstack001_r008_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='aNYMykHkxezUxs-07xgikw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='qHQpdWcFu7FzF6l50000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well01_mCherry_zstack001_r008_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='_eUmLccyLWPhCAZBQ8a3zA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='jVytS8AyAHmHkYR30000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_Alexa488_zstack001_r001_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='QFmofkisy-sibXAuKrpfRw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='cw4F6bUB9zuMthCY0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_DAPI_zstack001_r001_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='f4Vij96NOq7M96SXfyEYXQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='7TZGXvbA0JLL68hR0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_mCherry_zstack001_r001_c007.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='qUKQaesz-vfPUwljqPlIjA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='vCVbKkzz4CnJPPKF0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_Alexa488_zstack001_r001_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='WyabAqK4jeZtTtrQ9rvUeQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC), Artifact(uid='5kMhlcDNek4RMeQF0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_DAPI_zstack001_r001_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='eXaBPwLJJ--9SyGD7JR0ew', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='9BmbViqMmlVhpfS00000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well01_mCherry_zstack001_r001_c008.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='EeQ_zoM4j8waIP8wP-u0Hg', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='OS0wBE7bviIlW7qj0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_Alexa488_zstack001_r001_c002.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='RwasAYhkUEeTVBFrwCbKuw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='ixOpuSTsyrPXdYuA0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_DAPI_zstack001_r001_c002.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='diUYJPHgU_SvK0zTrrzUlw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='6uMjKAk1aYlAV7Cf0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_mCherry_zstack001_r001_c002.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='9K0N7dMe59K6zYUtjE74cA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='9ZVngbl0JUS0XdZ70000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_Alexa488_zstack001_r001_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='W1Q1RQCUj7j4e9R5Sta4wQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='IzP3IAwIhmM7OORD0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_DAPI_zstack001_r001_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='dIUw6okO0jOq6JcgsxDktA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='RRVS8qVx3VSw02Xu0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well01_mCherry_zstack001_r001_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='9ZmlWOdgh9GFPd4BCYO7GQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='Gtwi9Pcyx8maQEWB0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_Alexa488_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='yJMlFFYyup0JmBu_FjTa3g', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC), Artifact(uid='sHHpiiFYWsIXMZNV0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_DAPI_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='6pvuFREF3CQ8xr8Urak52Q', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='hNMkrIHce1XrLZHY0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_mCherry_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='K1jGiIE0dljR_qYP1btlyw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='nSZhAypqiNZ2Ylbe0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_Alexa488_zstack001_r002_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='uh537K7rMhMNKW-mti1vjw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='jzqxtoduIJ3hCbB40000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_DAPI_zstack001_r002_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='SOxM7EH80KPXK3hzBxXYLg', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC), Artifact(uid='M06liaIzh2OVEuJ40000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row01_Well08_mCherry_zstack001_r002_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='0tV4uQYxnP6G8JOrAwMyXw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='mXWwV1x42Jz9RoSO0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_Alexa488_zstack001_r002_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='4xc4bpAMo0UaM_tc8SciIA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='gj0HHnoVpEqbaUJb0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_DAPI_zstack001_r002_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='fZ4W2ePHagfGUGCcve98gQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='1XnEyqVt6UGXCTmV0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_mCherry_zstack001_r002_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='if3WCKH91_CY0ptIVYuLmA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='Ov8FnKzHMNY0XVJa0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_Alexa488_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='5cfc02GejBaoJ0h3OLWSdw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='Lwk8shsYe0V5bMgd0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_DAPI_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='CmAa139DH_lQ2Plcy4BxXw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='YuyVn060M4FxATPz0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row02_Well08_mCherry_zstack001_r002_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='gZHwk76xPz7qXiST4RuVMQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='ThuJnRAhqkp54kyU0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_Alexa488_zstack001_r010_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='vs3t8d40w259zgJRltHlKA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='PquYNyshQTDd24Vw0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_DAPI_zstack001_r010_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='1mSZ_MMg393bfTvMOl3YMQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='8SFUmW0RhBNySxBO0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_mCherry_zstack001_r010_c009.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='fjO8ly0euUPHXoQd86weww', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='h4EKWveW36LIzXez0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_Alexa488_zstack001_r010_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='ZPUWq_TI3KWNcVeVz3vOPg', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='fdem35nw5ztUnEIM0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_DAPI_zstack001_r010_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='XuugTTV9S5TbJKVMLOaR0w', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='VkmKLUCaMsYFCuGE0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row03_Well08_mCherry_zstack001_r010_c010.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='A-MzQvq-kHeu1WHdVmm5bw', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='QDPX1ljp0eCMz80o0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_Alexa488_zstack001_r001_c003.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='Y1bgVRJGzIEhKFGP54fplA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:24 UTC), Artifact(uid='Cvamog4G3a2XYGM80000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_DAPI_zstack001_r001_c003.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='UFjAP7uBiaM9ivE4hJzHTA', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='6uUjyphUD4D1Hixc0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_mCherry_zstack001_r001_c003.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='lvFoCpbAkjg4mEtXx-D99w', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
! cannot infer feature type of: <ArtifactQuerySet [Artifact(uid='Oww4y0yYuR8pxV9q0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_Alexa488_zstack001_r001_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='YX1aIPYndKFiPl5W3OPqfQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='AhBvnNKg5yJcG6LU0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_DAPI_zstack001_r001_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='9I6amjQc298AZvER1Z2jjQ', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC), Artifact(uid='AVRTVX9gEu4LrTAP0000', is_latest=True, key='input_data_imaging_usecase/images/Timepoint001_Row04_Well08_mCherry_zstack001_r001_c005.tif', description='raw image of U2OS cells stained for autophagy markers', suffix='.tif', size=2333056, hash='rYoK8xKXBtx5aNW4AfUbUg', branch_id=1, space_id=1, storage_id=2, run_id=2, created_by_id=1, created_at=2025-03-07 13:51:25 UTC)]>, returning '?
Dataset already processed. Skipping.
example_artifact = ln.Artifact.filter(
ulabels=ln.ULabel.get(name="scportrait single-cell images")
).first()
example_artifact.view_lineage()
ln.finish()
Show code cell output
→ finished Run('MaUzrpv5') after 1m at 2025-07-29 19:24:44 UTC

